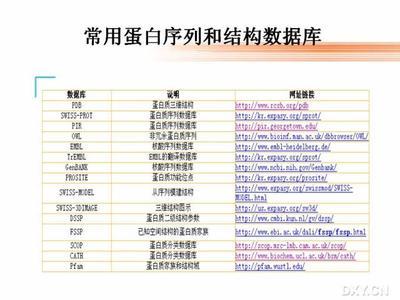
网上生物信息学教程 http://www.bio.cam.ac.uk/Embnetut/Gcg/index.html http://www.nal.usda.gov/pgdic 生物信息学机 http://www.ncbi.nlm.nih.gov/ EBI http://www.ebi.ac.uk/ http://www.nal.usda.gov/ http://genomic.sanger.ac.uk/ http://www.cbi.pku.edu.cn 数据库信息发布及其它 dbEST summary report http://www.ncbi.nlm.nih.gov/dbEST/dbESTsummarv.html Eukaryotic promoter database releasenotes http://www.genome.ad.jp/dbget/dbget2.html 核苷酸数据库 http://www.ncbi.nlm.nih.gov/ http://www.ncbi.nlm.nih.gov/dbEST/index.html http://www.ncbi.nlm.nih.gov/dbSTS/index.html dbGSS http://www.ncbi.nlm.nih.gov/dbGSS/index.html http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Genome http://www.ncbi.nlm.nih.gov/SNP/ http://www.ncbi.nlm.nih.gov/HTGS/ http://www.ncbi.nlm.nih.gov/UniGene/ http://www.ebi.ac.uk/embl http://www.ebi.ac.uk/genomes/ http://www.ebi.ac.uk/embl/Submission/webin.html http://www.ddbj.nig.ac.jp/ http://www.epd.isb-sib.ch/ http://www.gene-regulation.com/pub/databases.html TFD http://www.ifti.org http://www.geneontology.org 蛋白质数据库SWISS-PROT或TrEMBL http://www.ebi.ac.uk/swissprot/ PIR http://www-nbrf.georgetown.edu/pir/ http://www.prf.or.jp/ http://www.genome.ad.jp/dbget-bin/www_bfind?pdbstr Prosite http://www.expasy.ch/sprot/prosite.html http://www.rcsb.org/pdb http://www.pdb.org http://ndbserver.rutgers.edu/NDB/ndb.html http://ndbserver.rutgers.edu/ http://ndbserver.rutgers.edu/NDB/structure-finder/nmr/index.html http://ndbserver.rutgers.edu/NDB/structure-finder/protein/index.html http://www.expasy.ch/sw3d/ http://scop.mrc-lmb.cam.ac.uk/scop/ http://www.biochem.ucl.ac.uk/bsm/cath/ 酶、代谢和调控路径数据库KEGG http://www.genome.ad.jp/kegg/ http://www.genome.ad.jp/dbget/ligand.html http://www.cme.msu.edu/WIT/ EcoCyc http://ecocyc.PangeaSystems.com/ecocyc/ http://www.labmed.umn.edu/umbbd/ http://www.unl.edu/stc-95/ResTools/biotools/biotools8.html http://transfac.gbf.de http://www.gramene.org http://www.graingenes.org http://www.transgenica.com/botanicaldatabase.htm http://www.calflora.org/calflora/batanical.html http://rgp.dna.affrc.go.jp http://www.genome.clemson.edu/projects/rice/fpc http://www.genomics.org.cn http://www.genoscope.cns.fr http://www.agron.missouri.edu http://www.css.orst.edu/Research/barley/nabgmp.htm http://forages.orst.edu/ http://wheat.pw.usda.gov/index.shtml http://www.arabidopsis.org http://129.186.26.94 http://www.alfalfa.ksu.edu http://cottongenomecenter.ucdavis.edu http://www.zmdb.iastate.edu/PlantGDB/glycine_max.html genome http://www.acedb.org http://www.biology.duke.edu/chlamy_genome http://dictygenome.bcm.tmc.edu http://www.thearkdb.org http://flybase.bio.indiana.edu/ http://www.informatics.jax.org/ Saccharomyces GenomeDatabase |
多种基因组数据库 http://www.hgmp.mrc.ac.uk/GenomeWeb http://www.ncbi.nlm.nih.gov/PubMed/ http://www.ncbi.nlm.nih.gov/Omim/ http://www.nal.usda.gov/ag98/ http://www.gramene.org/newsletters/————rice_genetics http://www.ncbi.nlm.nih.gov/Entrez/ Sequence Retrieval System,Singapore http://iubio.bio.indiana.edu:80/srs/srsc http://srs.ebi.ac.uk/ http://ftp2.ddbj.nig.ac.jp:8000/getstart-e.html http://ftp2.ddbj.nig.ac.jp:8080/dbsearch-e-new.html http://www.genome.ad.jp/dbget/dbget2.html http://www.ncbi.nlm.nih.gov/BLAST/ http://www.ebi.ac.uk/fasta33/index.html http://www2.ebi.ac.uk/bicsw/
http://www2.igh.cnrs.fr/bin/ssearch-guess.cgi http://www.ncbi.nlm.nih.gov/STS/ http://www.ebi.ac.uk/proteome/ http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html http://www.ebi.ac.uk http://onyx.si.edu/PAUP/ http://www.ebi.ac.uk http://www.gcg.com/ http://evolution.genetics.washington.edu/phylip.html http://www.bio.psu.edu/imeg http://www.vims.edu/~mes/hennig/software.html http://www.lifesci.ucla.edu/mcdbio/Faculty/Lake/Research/Programs/ http://phylogeny.arizona.edu/macclade/macclade.html http://www.unl.edu/stc-95/ResTools/biotools/biotools2.html ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX http://www.megasoftware.net http://genes.mit.edu/GENSCAN.html http://genomic.sanger.ac.uk/gf/gf.shtml http://dot.imgen.bcm.tmc.edu:9331/ http://compbio.ornl.gov/Grail-1.3/ http://grail.lsd.ornl.gov/grailexp/ http://opal.biology.gatech.edu/GeneMark/eukhmm.cgi http://www.cs.jhu.edu/labs/compbio/veil.html
http://genome.cs.mtu.edu/aat.html http://www.imim.es/GeneIdentification/Geneid/geneid_input.html http://cbil.humgen.upenn.edu/~sdong/genlang_home.html GeneParser http://beagle.colorado.edu/~eesnyder/GeneParser.html http://www.cs.jhu.edu/labs/compbio/glimmer.html http://www.cshl.org/genefinder http://www-hto.usc.edu/software/procrustes/
http://udgenome.ags.udel.edu/gofigure http://www.expasy.ch/ http://www.cbs.dtu.dk http://dove.embl-heidelberg.de/3D/ http://www.bork.embl-heidelberg.de/Frame/ MatInspector http://www.gsf.de/cgi-bin/matsearch.pl http://www.gsf.de/cgi-bin/fastm.pl http://www.dna.affrc.go.jp/htdocs/sigscan/signal.html ProDom 其它 多种数据库、分析工具和生物信息学机构 |
 爱华网
爱华网